001、
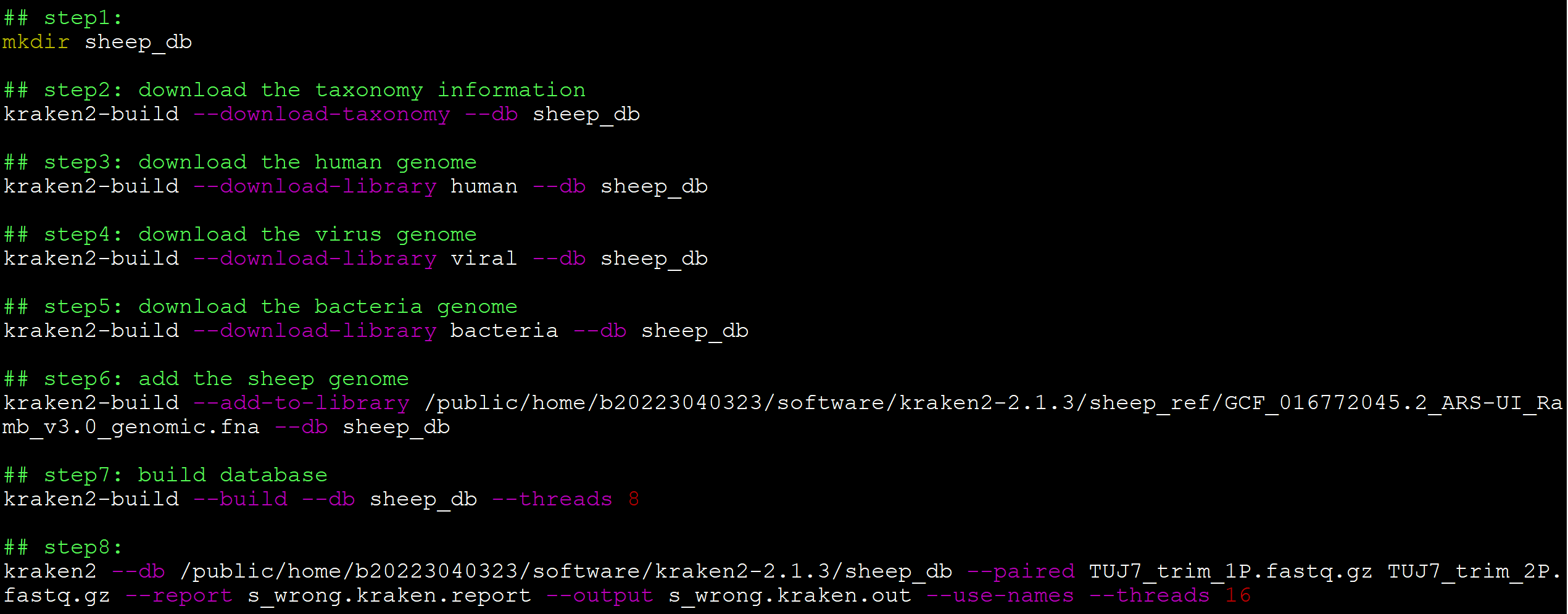
## step1: mkdir sheep_db ## 创建数据库目录## step2: download the taxonomy information kraken2-build --download-taxonomy --db sheep_db ## 下载物种分类信息## step3: download the human genome kraken2-build --download-library human --db sheep_db ## 下载人类基因组## step4: download the virus genome kraken2-build --download-library viral --db sheep_db ## 下载病毒基因组## step5: download the bacteria genome kraken2-build --download-library bacteria --db sheep_db ## 下载细菌基因组## step6: add the sheep genome kraken2-build --add-to-library /public/home/b20223040323/software/kraken2-2.1.3/sheep_ref/GCF_016772045.2_ARS-UI_Ramb_v3.0_genomic.fna --db sheep_db ## 添加绵羊基因组## step7: build database kraken2-build --build --db sheep_db --threads 8 ## 构建数据库## step8: kraken2 --db /public/home/b20223040323/software/kraken2-2.1.3/sheep_db --paired TUJ7_trim_1P.fastq.gz TUJ7_trim_2P.fastq.gz --report s_wrong.kraken.report --output s_wrong.kraken.out --use-names --threads 16 ## 运算
。
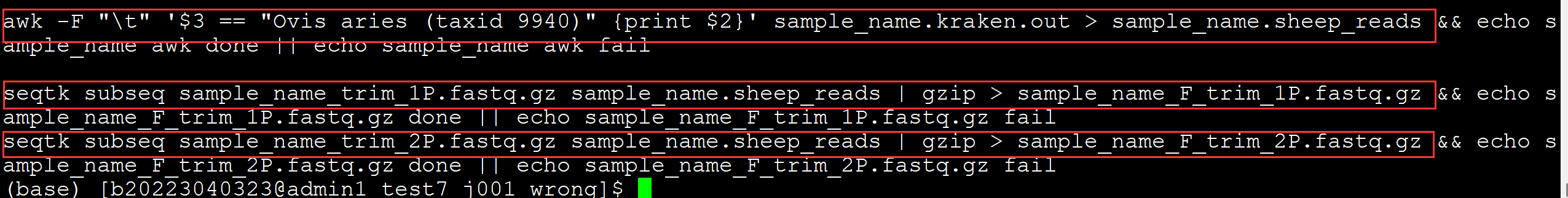
002、 提取目标物种的reads
awk -F "\t" '$3 == "Ovis aries (taxid 9940)" {print $2}' sample_name.kraken.out > sample_name.sheep_reads ## 提取reads 的 IDseqtk subseq sample_name_trim_1P.fastq.gz sample_name.sheep_reads | gzip > sample_name_F_trim_1P.fastq.gz ## 提取read1 seqtk subseq sample_name_trim_2P.fastq.gz sample_name.sheep_reads | gzip > sample_name_F_trim_2P.fastq.gz ## 提取read2
。
